
Red Wine
Fermented alcoholic grape beverage
Ingredients: grapes
Taste: varying in flavor, tannic, sweet, alcoholic
Popular in: global
Time: 3 months+
Dominant microbes: Yeast and lactic acid producers
Tools: large vessel, bottles
red wi·ne
ˈred /ˈwīn /
ˈred /ˈwīn /
ˈred /ˈwīn /
Recipe:
Basic Grape Red Wine
Ingredients
grapes
Tools
large bowl or
crock (non-metal)
wooden spoon
cheesecloth
carboy or clean glass jug
sterile funnel and sieve
airlock
- Place the grapes in a large clean bowl or bucket (depending on the amount), crush or press the grapes to get out as much juice as possible. Removing the stems is optional (adds more tannins).
- Once all the juice has been squeezed out, stir vigorously, and cover with a cheese cloth to allow the grapes to breath but prevent flies; leave at room temperature.
- Stir vigorously 3-4 times a day to prevent mold from growing on the surface
and wipe down the sides of the jar keeping it clean. Do this for 2-3 days,
or until you begin to see really active bubbles from the active fermentation. - At this point, transfer to a carboy or glass jug to begin secondary fermentation. Using the sieve to strain out any stems or extra skins (you can also leave them in), funnel the wine into the carboy or glass jug, and close with an airlock.
- Store in a cool, dark place like a basement or closet, around 55-60°F
No airlock? No problem.
If you don’t have an airlock, you can cap the jug or place an empty balloon on top. However, it is important to open the cap everyday (or twice a day) for the first one-two weeks to ‘burp’ the wine, otherwise the CO2 will build up and the bottle will explode. By placing a balloon on the top, you can see how much CO2 builds up as it expands.
wine microbiology
who are the key players?
YEAST - 1° players in alcohol fermentation
Saccharomyces cerevisiae dominates the final ferment; there are as many as 100 distinct strains of S. cerevisiae (Parish and Fleet, 2012). While most strains of S. cerevisiae will produce a reliably similar amount of alcohol, carbon dioxide, and other primary metabolites, specific strains will have a tremendous impact on the final flavor profile. “Seeded” fermentation, or wine that uses a commercial yeast starter to kick off the fermentation process uses S. cerevisiae strains selected for a specific and consistent flavor profile.
Other non-S. cerevisiae yeast isolated include: Kloeckera apiculata, Candida stellata, Candida colliculosa, Candida pulcherrima, and Hansenula anomala.
With some exception, S. cerevisiae will eventually dominate the primary fermentation of the wine, specifically suppressing certain non-S. cerevisiae yeasts in both natural and inoculated wine ferments.
Inoculation with commercial yeast does not mean wild yeast will not be present. Even following addition of sulfur dioxide at 50mg/L and inoculating the grape must with commercial S. cerevisiae at 105 to 107 cells showed that although the S. cerevisiae dominated the wine, other yeasts still grew, contributing to the final composition of the wine (Fleet, 1993). Additionally, there remains the possibility that the strain of S. cerevisiae that dominates the final wine is a wild strain, not the inoculated strain* (Heard and Fleet, 1985). Mycobiome dynamics in wine making remain to be explored (Setati et al. 1988).
Certain yeast are also primarily associated with spoileage, including Pichia mebranifaciens, Zygosaccharomyces bailii, and Saccharomyces ludwigii, as well as members of the Brettanomyces and Dekkera genus.
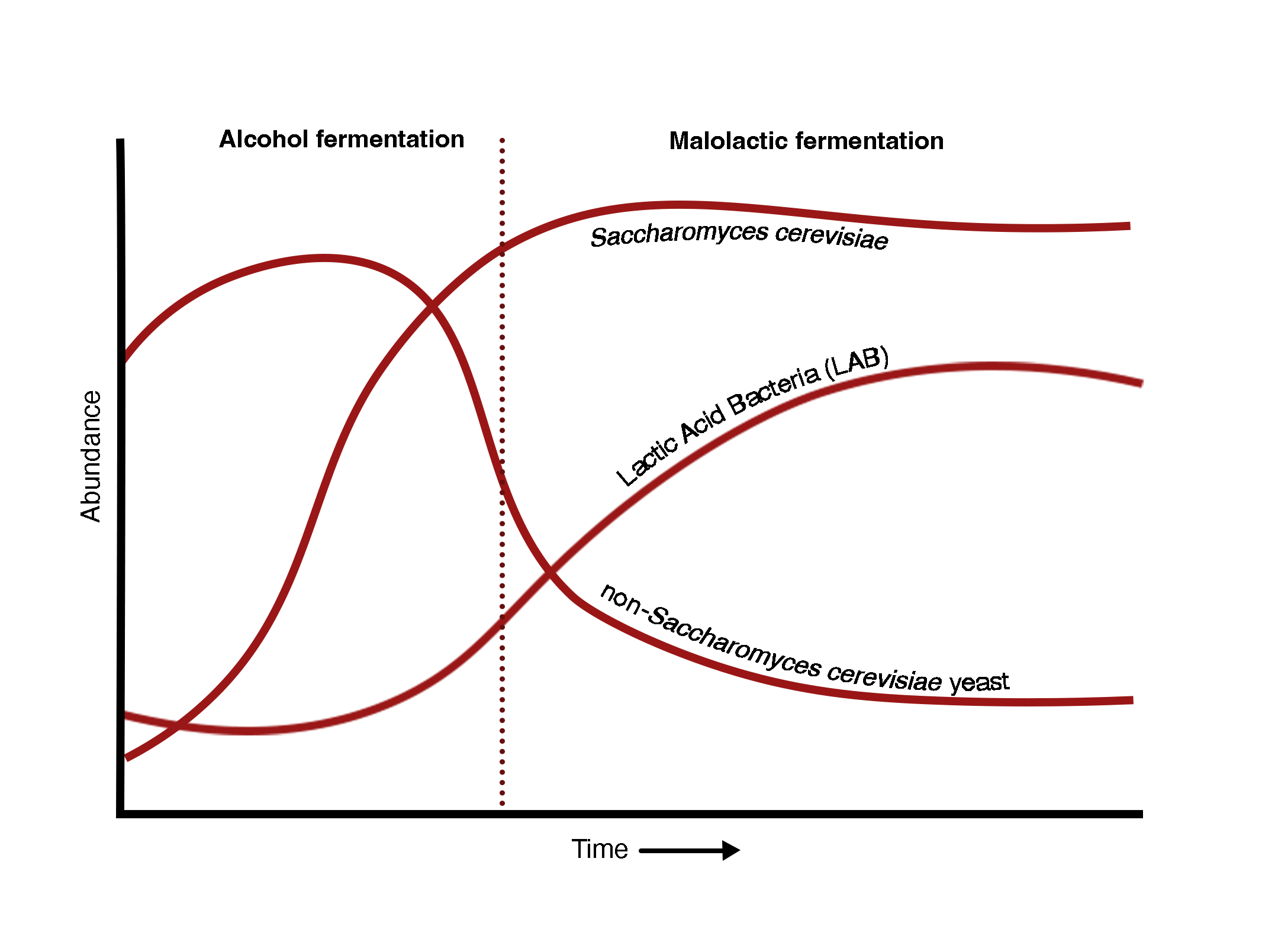
BACTERIA - 1° players in malolactic fermentation
Bacteria play an important role in malolactic fermentation (or secondary fermentation), which occurs after the initial alcohol fermentation.
Lactic acid bacteria (LAB), such as Lactobacillus, Leuconostoc, Oenococcus and Pediococcus have been identified as playing a role. Oenococcus oeni is considered the preferred species for favorable malolactic fermentation, and can be added in commercial wine making at the beginning of malolactic fermentation. Other genuses like Lactobacillus and Pediococcus can lead to the formation of off flavors.
Factors that might prevent LAB growth (and thus malolactic fermentation) in wine:
-
ph < 3
- ethanol > 12% v/v
- Sulfur dioxide > 50ug/mL
The impact of these factors on strain growth is very strain dependent; Generally, as a species Oenococcus oeni seems to be more tolerant of a lower pH and less tolerant to high sulfur dioxide compared to other LAB.
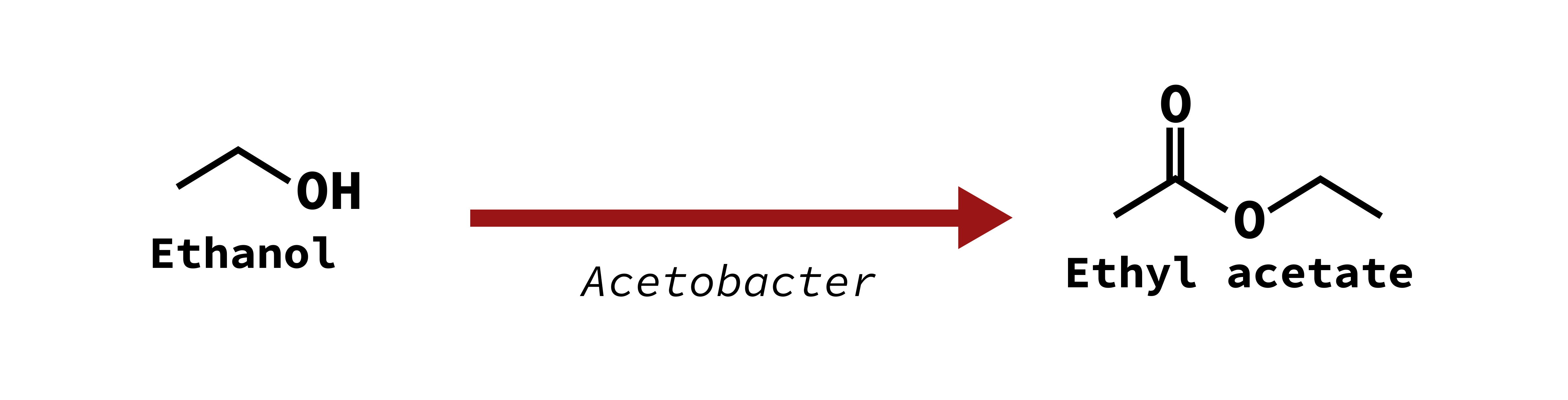
All acetic acid bacteria species (including Acetobacter and Gluconobacter) are considered spoilage bacteria, as they can metabolize ethanol into acetic acid and produce a vinegar flavor or ethyl acetate producing a nail polish remover-like scent. AAB can be found in higher quantities on spoiled grapes, where they end up growing with the yeast during the primary fermentation step and drastically impact flavor. On healthy, undamaged grapes they are found in low enough abundance that AAB does not seem to contribute to final flavor.
alcohol fermentation
the primary fermentation stage in wine making, usually takes place in the first week following crushing of the grapes, and is complete sugars have been utilized; at this point, the majority of alcohol has been produced and S. cerevisiae is the dominant microbe.
mycobiome
from the Greek mukēs or “fungus” and bíos or “life”, this refers to the fungal community in or on an organism
the primary fermentation stage in wine making, usually takes place in the first week following crushing of the grapes, and is complete sugars have been utilized; at this point, the majority of alcohol has been produced and S. cerevisiae is the dominant microbe.
mycobiome
from the Greek mukēs or “fungus” and bíos or “life”, this refers to the fungal community in or on an organism
*if anyone knows of any studies looking at inoculated S. cerevisiae strain tracking over the course of fermentation, let me know.
malolactic fermentation
the second fermentation stage in wine making. Following alcohol fermentation, wine is transferred to barrels or stainless steel tanks (a low oxygen environment) where lactic acid bacteria covert malic acid produced by yeast to lactic acid, decreasing the acidity. Malolactic fermentation is not necessary for all wines, but there is evidence that successfull malolactic fermentation inhibits growth of wine spoileage organisms like Brettanomyces.IMMATURE GRAPES
MATURE GRAPES
Only 10-1000 CFU/g yeast cells are found
on immature grapes
Primary yeast on immature grapes:
Rhodotorula sp.
Sporobolomyces sp.
Crytococcus sp.
Candida sp.
on immature grapes
Primary yeast on immature grapes:
Rhodotorula sp.
Sporobolomyces sp.
Crytococcus sp.
Candida sp.
By the time the grapes are ready for harvest, about 10,000-1,000,000 CFU/g of yeast cells are found on the surface of the grapes, primarily non-Saccharomyces
Primary yeast on mature grapes:
Kloeckera apiculata
Candida stellata
Candida colliculosa
Candida pulcherrima
Hansenula anomala
Primary yeast on mature grapes:
Kloeckera apiculata
Candida stellata
Candida colliculosa
Candida pulcherrima
Hansenula anomala
If there is little yeast on the surface of the immature grape, how does it get there by the time of harvest?
Climate, soil, terrain, and grape variety are key participant in the development of the vineyard soil microbiome, hypothesized to be a reservoir for the wine fermentation microbiome. There are a few methods of dispersal from the soil to the grape that have been proposed:
- Physical contact through rain, dust, and wind
- Migration through the rhizosphere
- Insects moving from soil to fruits, or fruit to fruit
Additionally, grape variety might play a gatekeeping role in which microbes can establish themselves on the surface, based on the macronutient and micronutrients of the surface, and the climate best suited for the plant.
However, microbes identified on the grape surface are not the primary microbes found during wine fermentation-- how that community is reshaped reamins to be understood (Ramírez et al. 2020).
Soil and must fungal diversity are the main predictors of wine regionality, over climate, soil properties, and bacterial diversity (Liu et al. 2020)
If the grape surface is dominated by non-Saccharomyces yeast at harvest, how does S. cerevisiae become the dominant yeast species by the end of wine fermentation?
No identifcation of S. cerevisiae on the grapes at harvest might not mean it isn’t present. It’s probably below the limit of detection of current identification methods.
This is a similar problem with sauerkraut-- lactic acid bacteria that drive the fermentation process are in very low abundance on the cabbage itself, but blossom and dominate once they have access to the carbohydrates in the plant (Miller et al. 2019).
S. cerevisiae, present but in very low abundance, might only begin to establish itself once maceration has taken place, where it has easy access to the nutrients and sugars in the grapes. S. cerevisiae could also be sourced from the winemaking equiptment, carried over from previous ferments and present in the winery air. While strain tracking of S. cerevisiae needs to be further addressed, it is likely a combination of the two.
Of course, seeding with S. cerevisiae using a starter will quickly allow for it to establish irself as the dominant strain.
This is a similar problem with sauerkraut-- lactic acid bacteria that drive the fermentation process are in very low abundance on the cabbage itself, but blossom and dominate once they have access to the carbohydrates in the plant (Miller et al. 2019).
S. cerevisiae, present but in very low abundance, might only begin to establish itself once maceration has taken place, where it has easy access to the nutrients and sugars in the grapes. S. cerevisiae could also be sourced from the winemaking equiptment, carried over from previous ferments and present in the winery air. While strain tracking of S. cerevisiae needs to be further addressed, it is likely a combination of the two.
Of course, seeding with S. cerevisiae using a starter will quickly allow for it to establish irself as the dominant strain.
paths to spoileage
While there are several hundreds of metabolites known to contribute to the final flavor profile of the wine, there are also metabolites known to play a role in off-flavors or spoileage.
SPOILAGE FLAVOR
METABOLITEs RESPONSIBLE
POSSIBLE PATHWAYs and GENUS INVOLVED
Geranium notes
2-ethoxyhexa-3,5-dienne

Vinegar aroma
Acetic acid

Solvent character (nail polish remover)
Ethyl acetate
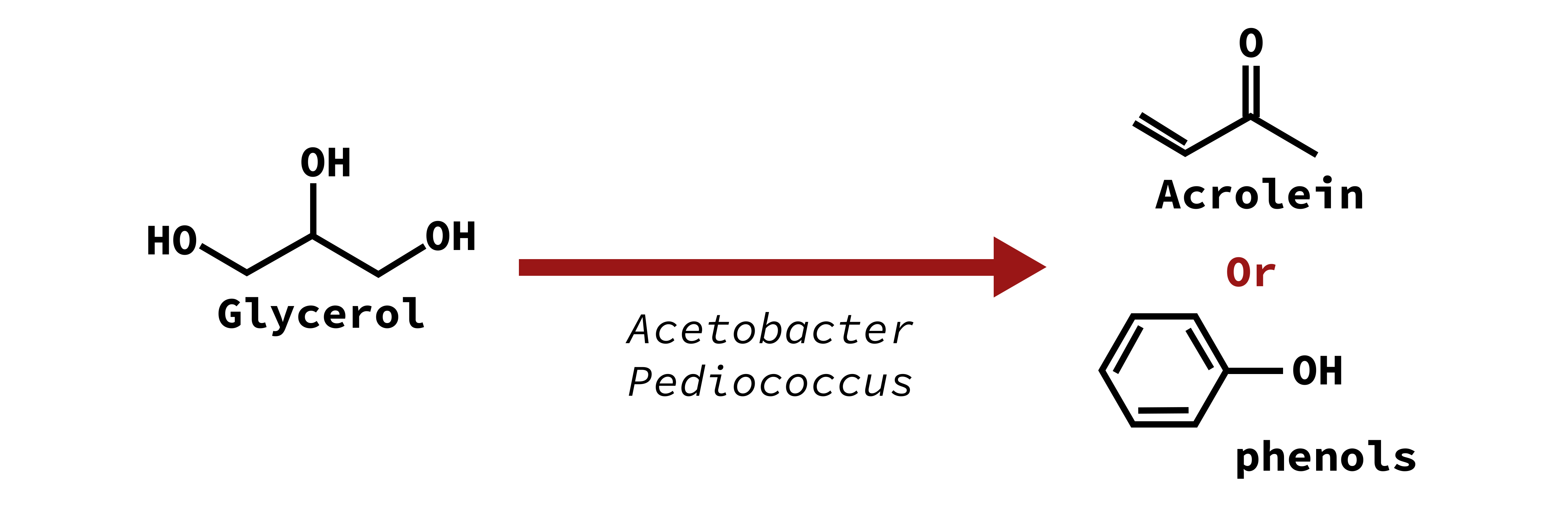
Bitterness
Acrolein and phenols
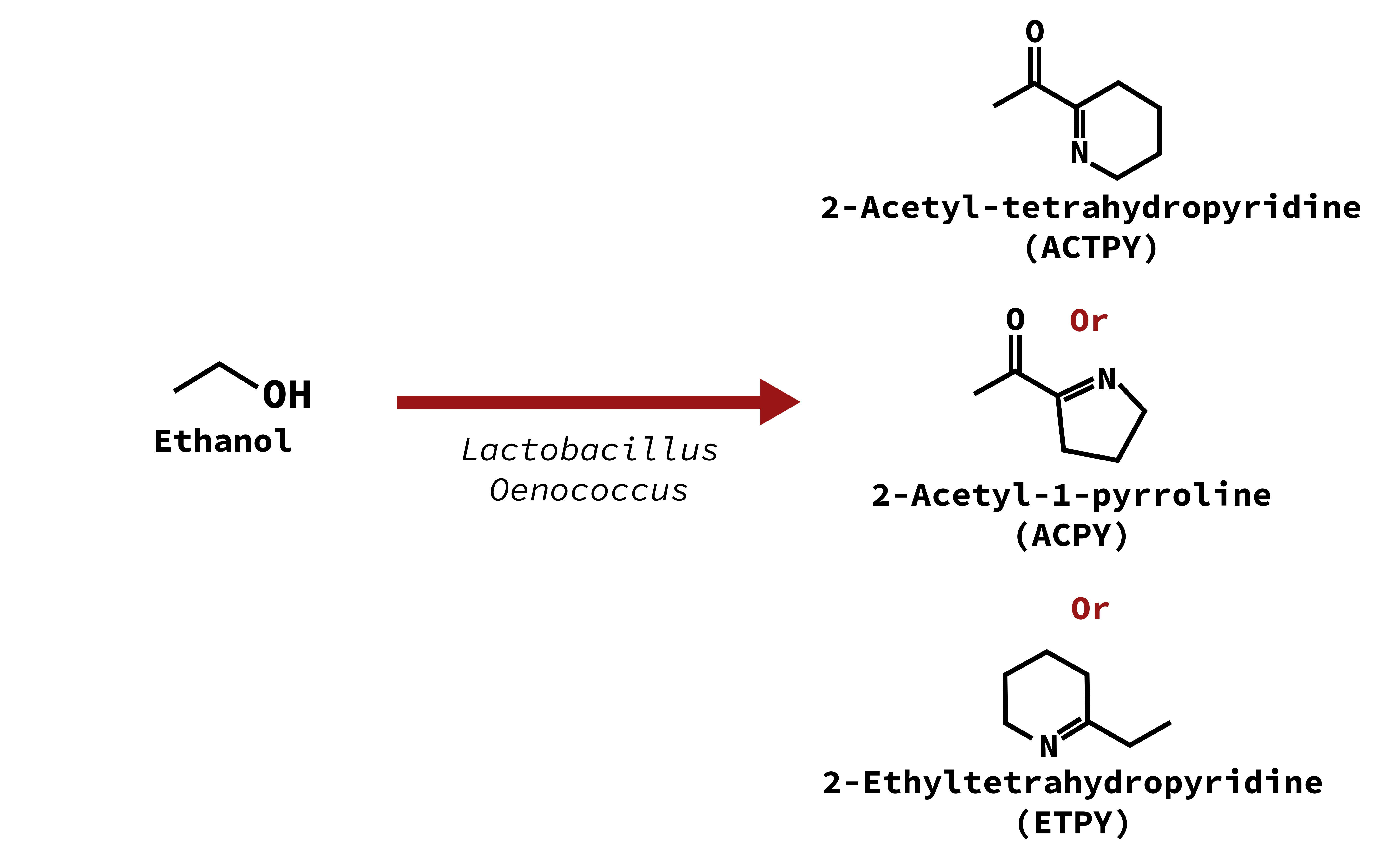
Mouse
2-Acetyl-tetrahydropyridine (ACTPY) 2-Ethyltetrahydropyridine (ETPY)
2-Acetyl-1-pyrroline (ACPY)
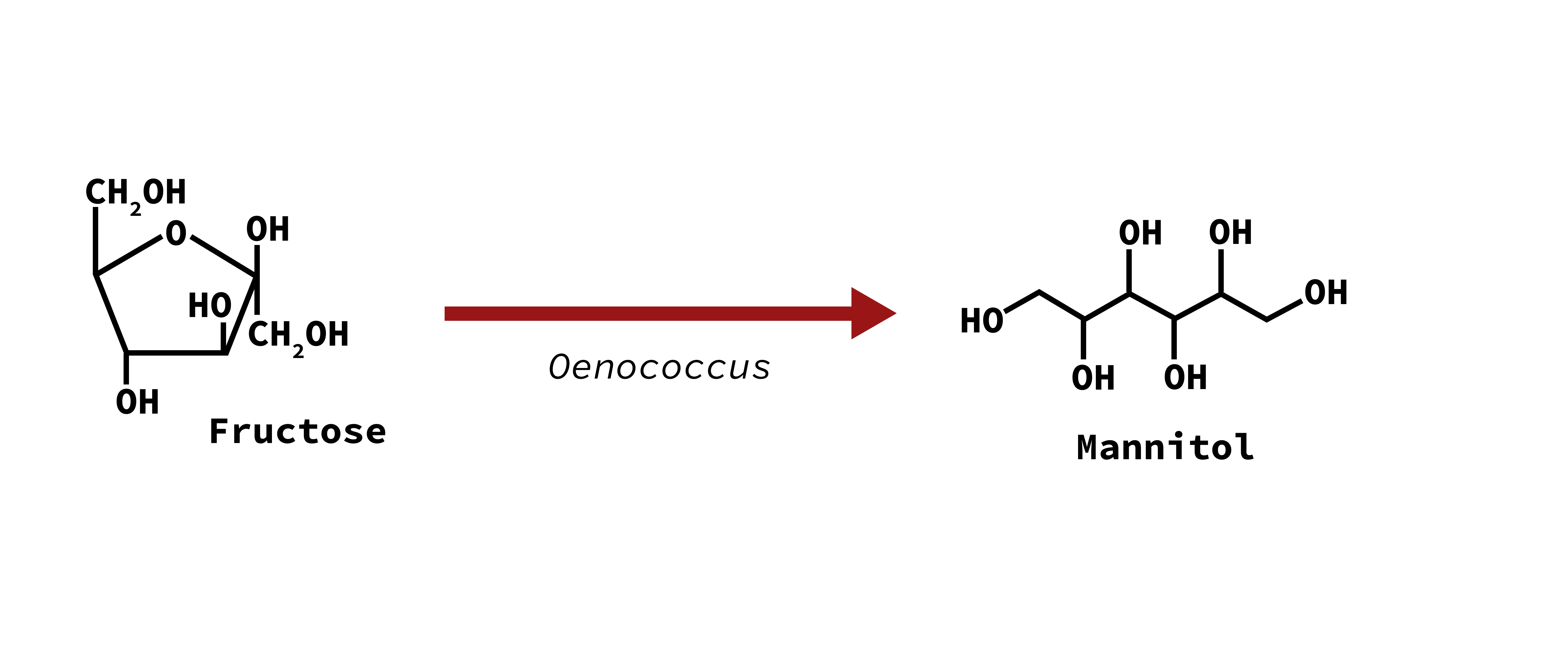
Viscous, irrirating finish (mannitol taint)
Mannitol
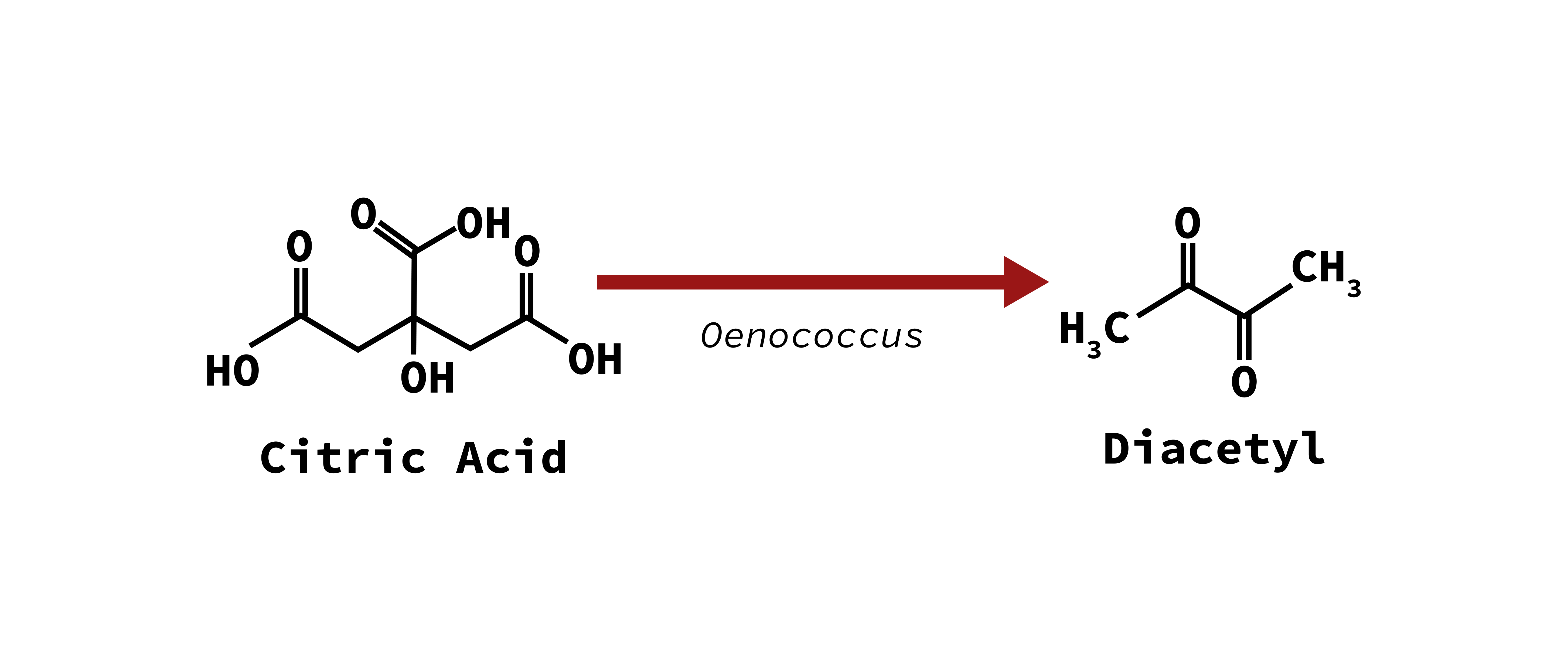
Buttery character
2,3 butandione (Diacetyl)
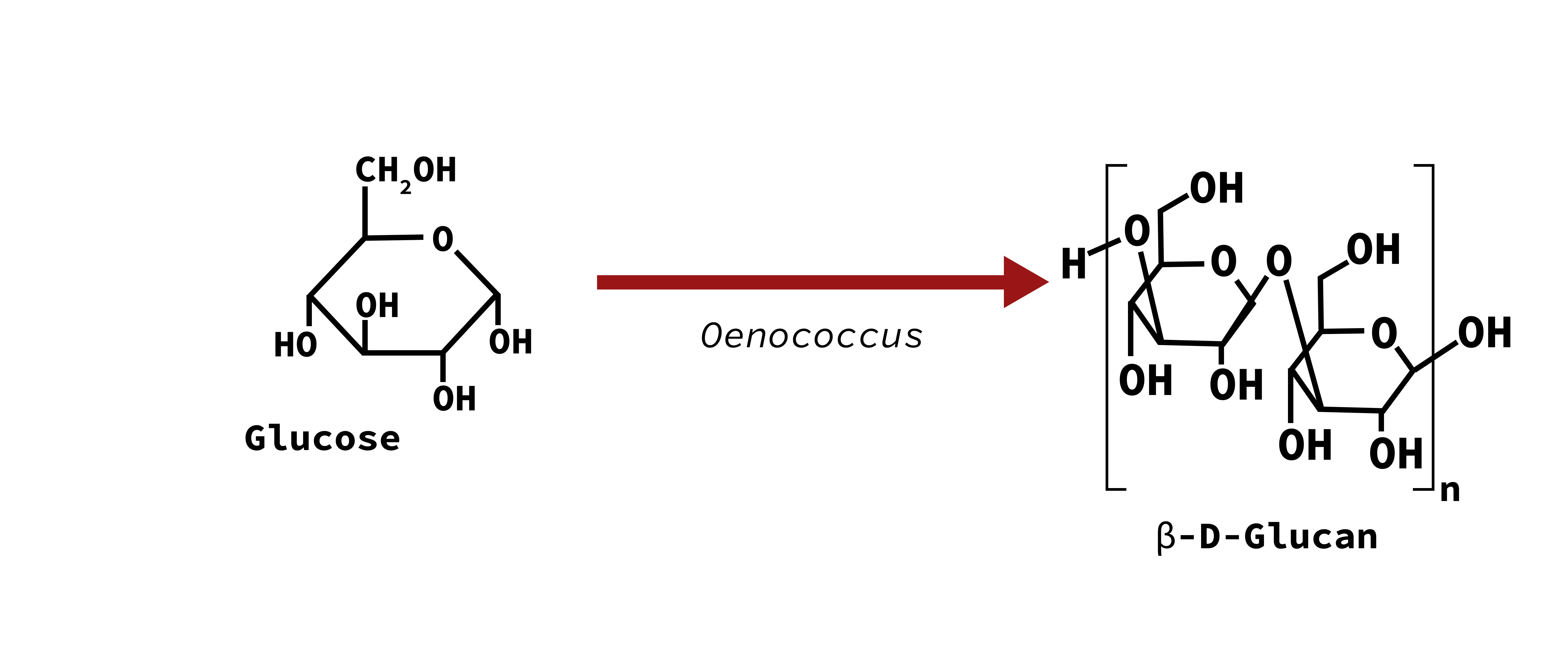
Ropy
β-D-Glucan
adapted from: Bartowsky, Evaline J. "Bacterial spoilage of wine and approaches to minimize it." Letters in applied microbiology 48.2 (2009): 149-156.S
additional reading
Bagheri, Bahareh, Florian F. Bauer, and Mathabatha E. Setati. "The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations." Frontiers in microbiology 8 (2017): 1988.
Bokulich, Nicholas A., et al. "Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics." MBio 7.3 (2016): e00631-16.
Fleet, Graham H. Wine microbiology and biotechnology. CRC Press, 1993.
Heard, Gillian M., and Graham H. Fleet. "Growth of natural yeast flora during the fermentation of inoculated wines." Applied and Environmental Microbiology 50.3 (1985): 727-728.
Liu, Di, et al. "The fungal microbiome is an important component of vineyard ecosystems and correlates with regional distinctiveness of wine." Msphere 5.4 (2020): e00534-20.
Parish, Mickey E., and Graham H. Fleet. "Wine." Food Microbiology: Fundamentals and Frontiers (2012): 915-947.
Ramirez, Manuel, et al. "Analysing the vineyard soil as a natural reservoir for wine yeasts." Food Research International 129 (2020): 108845.
Solieri, Lisa, and Paolo Giudici. "Development of a sequence-characterized amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation." Applied and environmental microbiology 76.23 (2010): 7765-7774.